Chapter 12 Bach mouse mammary gland (10X Genomics)
12.1 Introduction
This performs an analysis of the Bach et al. (2017) 10X Genomics dataset, from which we will consider a single sample of epithelial cells from the mouse mammary gland during gestation.
12.2 Data loading
12.3 Quality control
is.mito <- rowData(sce.mam)$SEQNAME == "MT"
stats <- perCellQCMetrics(sce.mam, subsets=list(Mito=which(is.mito)))
qc <- quickPerCellQC(stats, percent_subsets="subsets_Mito_percent")
sce.mam <- sce.mam[,!qc$discard]colData(unfiltered) <- cbind(colData(unfiltered), stats)
unfiltered$discard <- qc$discard
gridExtra::grid.arrange(
plotColData(unfiltered, y="sum", colour_by="discard") +
scale_y_log10() + ggtitle("Total count"),
plotColData(unfiltered, y="detected", colour_by="discard") +
scale_y_log10() + ggtitle("Detected features"),
plotColData(unfiltered, y="subsets_Mito_percent",
colour_by="discard") + ggtitle("Mito percent"),
ncol=2
)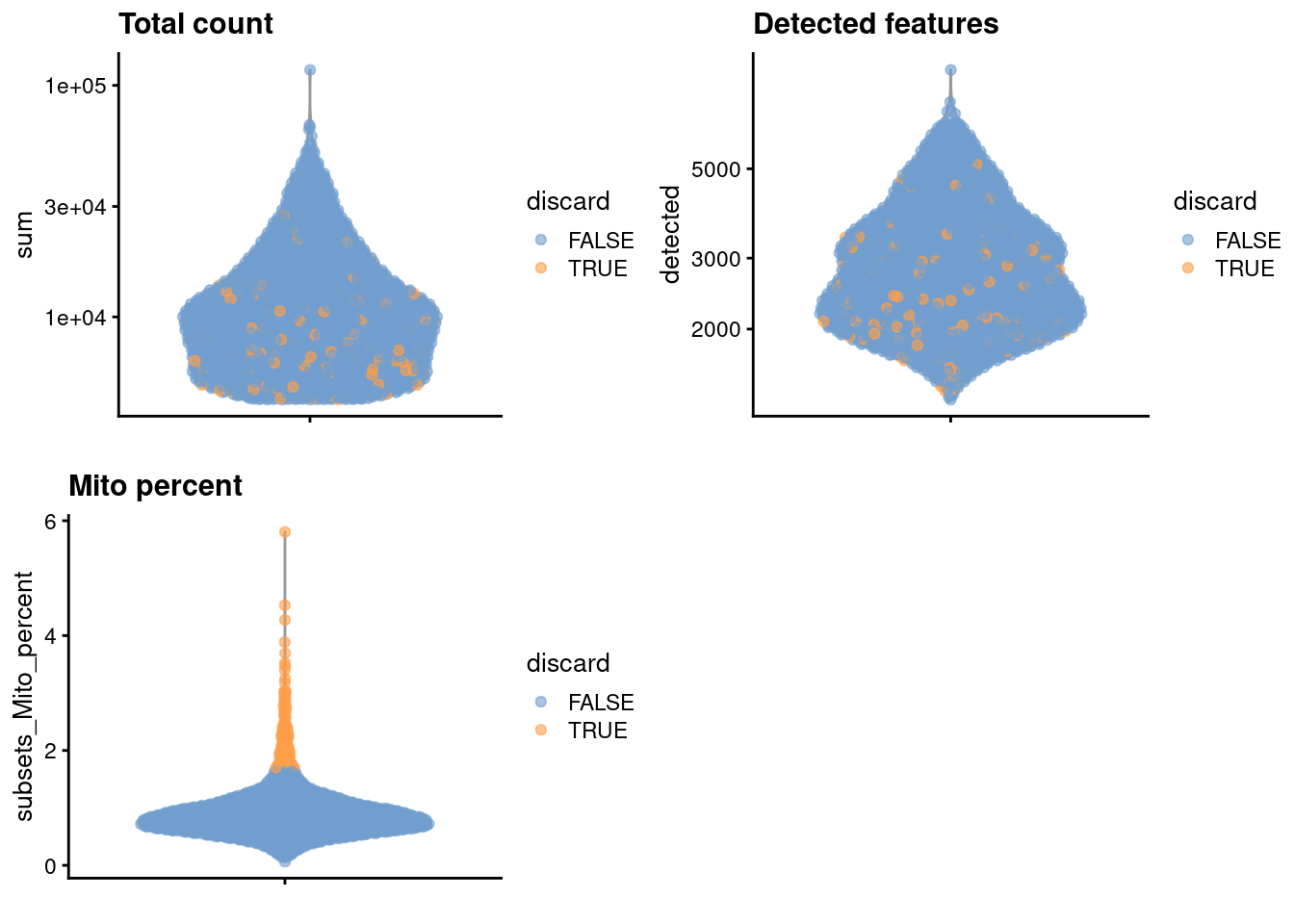
Figure 12.1: Distribution of each QC metric across cells in the Bach mammary gland dataset. Each point represents a cell and is colored according to whether that cell was discarded.
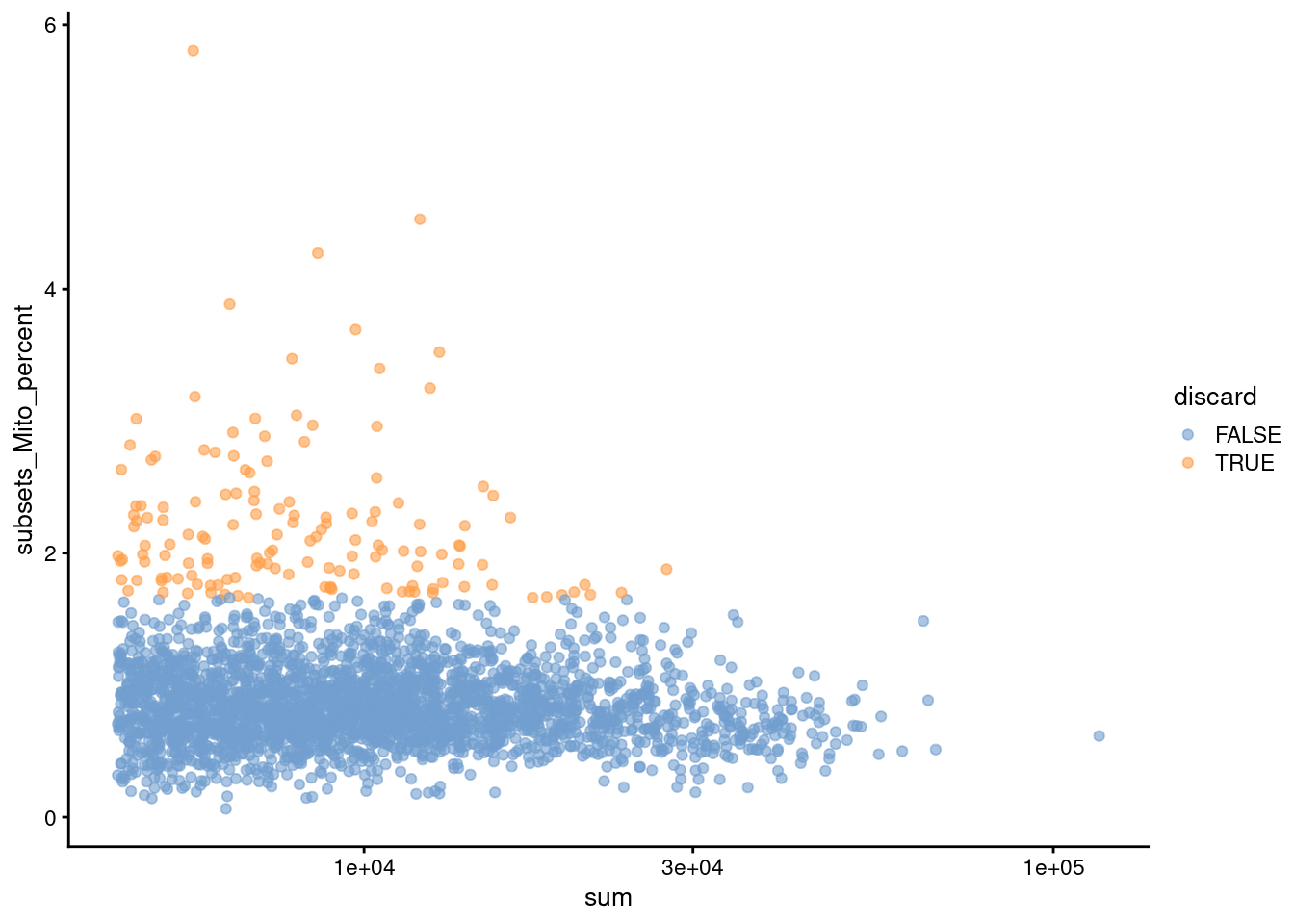
Figure 12.2: Percentage of mitochondrial reads in each cell in the Bach mammary gland dataset compared to its total count. Each point represents a cell and is colored according to whether that cell was discarded.
## low_lib_size low_n_features high_subsets_Mito_percent
## 0 0 143
## discard
## 14312.4 Normalization
library(scran)
set.seed(101000110)
clusters <- quickCluster(sce.mam)
sce.mam <- computeSumFactors(sce.mam, clusters=clusters)
sce.mam <- logNormCounts(sce.mam)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.271 0.522 0.758 1.000 1.204 10.958plot(librarySizeFactors(sce.mam), sizeFactors(sce.mam), pch=16,
xlab="Library size factors", ylab="Deconvolution factors", log="xy")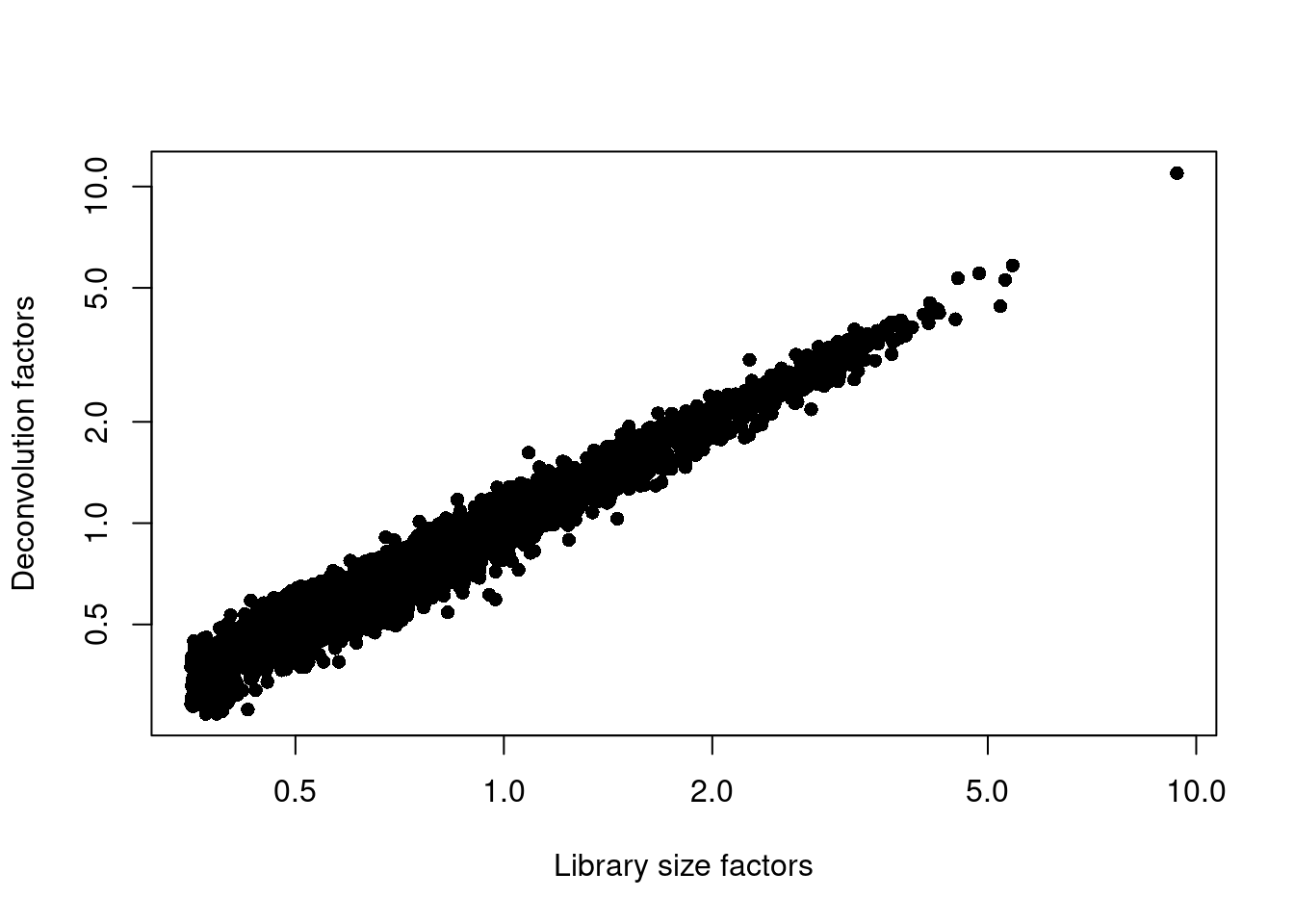
Figure 12.3: Relationship between the library size factors and the deconvolution size factors in the Bach mammary gland dataset.
12.5 Variance modelling
We use a Poisson-based technical trend to capture more genuine biological variation in the biological component.
set.seed(00010101)
dec.mam <- modelGeneVarByPoisson(sce.mam)
top.mam <- getTopHVGs(dec.mam, prop=0.1)plot(dec.mam$mean, dec.mam$total, pch=16, cex=0.5,
xlab="Mean of log-expression", ylab="Variance of log-expression")
curfit <- metadata(dec.mam)
curve(curfit$trend(x), col='dodgerblue', add=TRUE, lwd=2)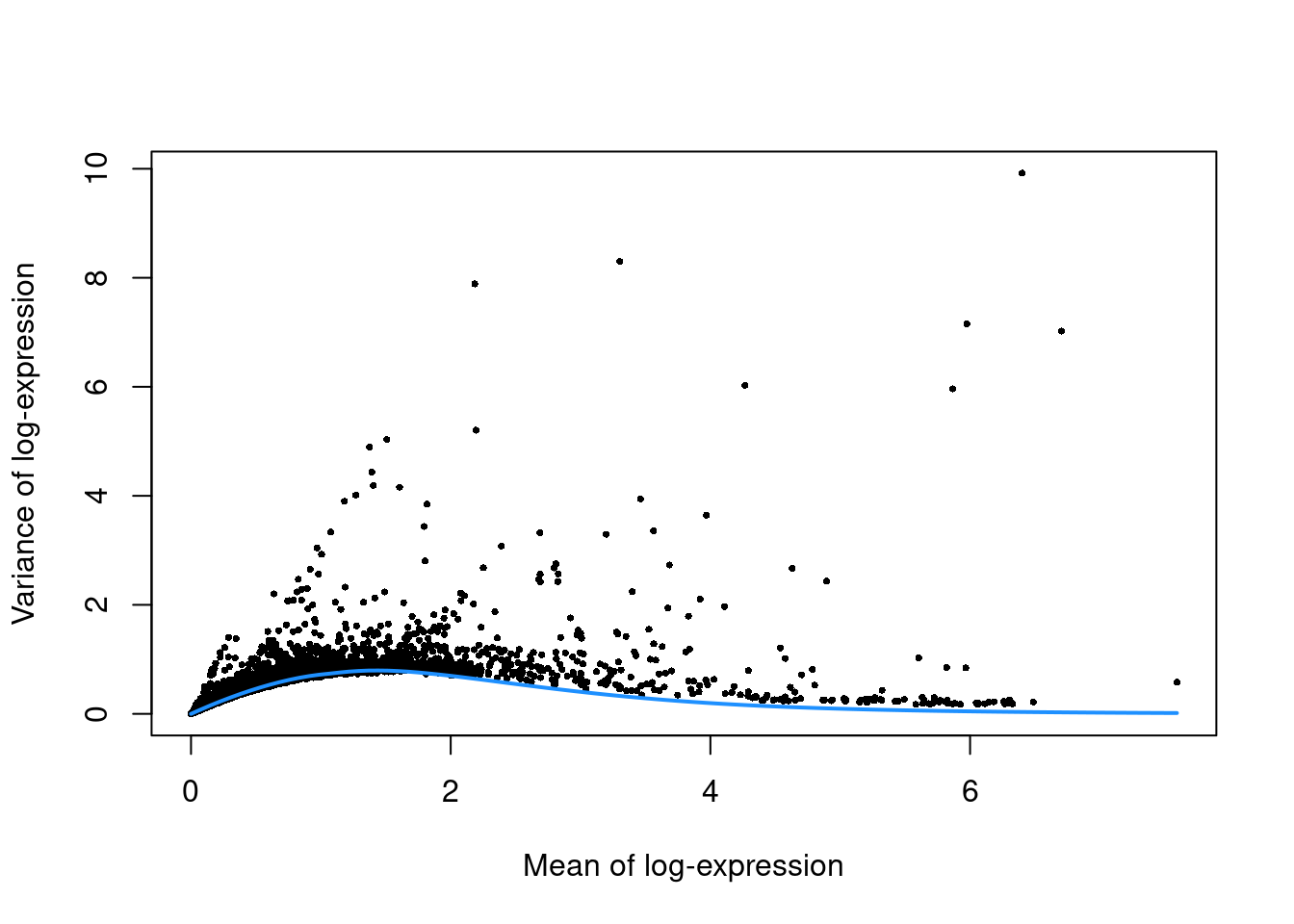
Figure 12.4: Per-gene variance as a function of the mean for the log-expression values in the Bach mammary gland dataset. Each point represents a gene (black) with the mean-variance trend (blue) fitted to simulated Poisson counts.
12.6 Dimensionality reduction
library(BiocSingular)
set.seed(101010011)
sce.mam <- denoisePCA(sce.mam, technical=dec.mam, subset.row=top.mam)
sce.mam <- runTSNE(sce.mam, dimred="PCA")## [1] 1512.7 Clustering
We use a higher k to obtain coarser clusters (for use in doubletCluster() later).
snn.gr <- buildSNNGraph(sce.mam, use.dimred="PCA", k=25)
colLabels(sce.mam) <- factor(igraph::cluster_walktrap(snn.gr)$membership)##
## 1 2 3 4 5 6 7 8 9 10
## 550 799 716 452 24 84 52 39 32 24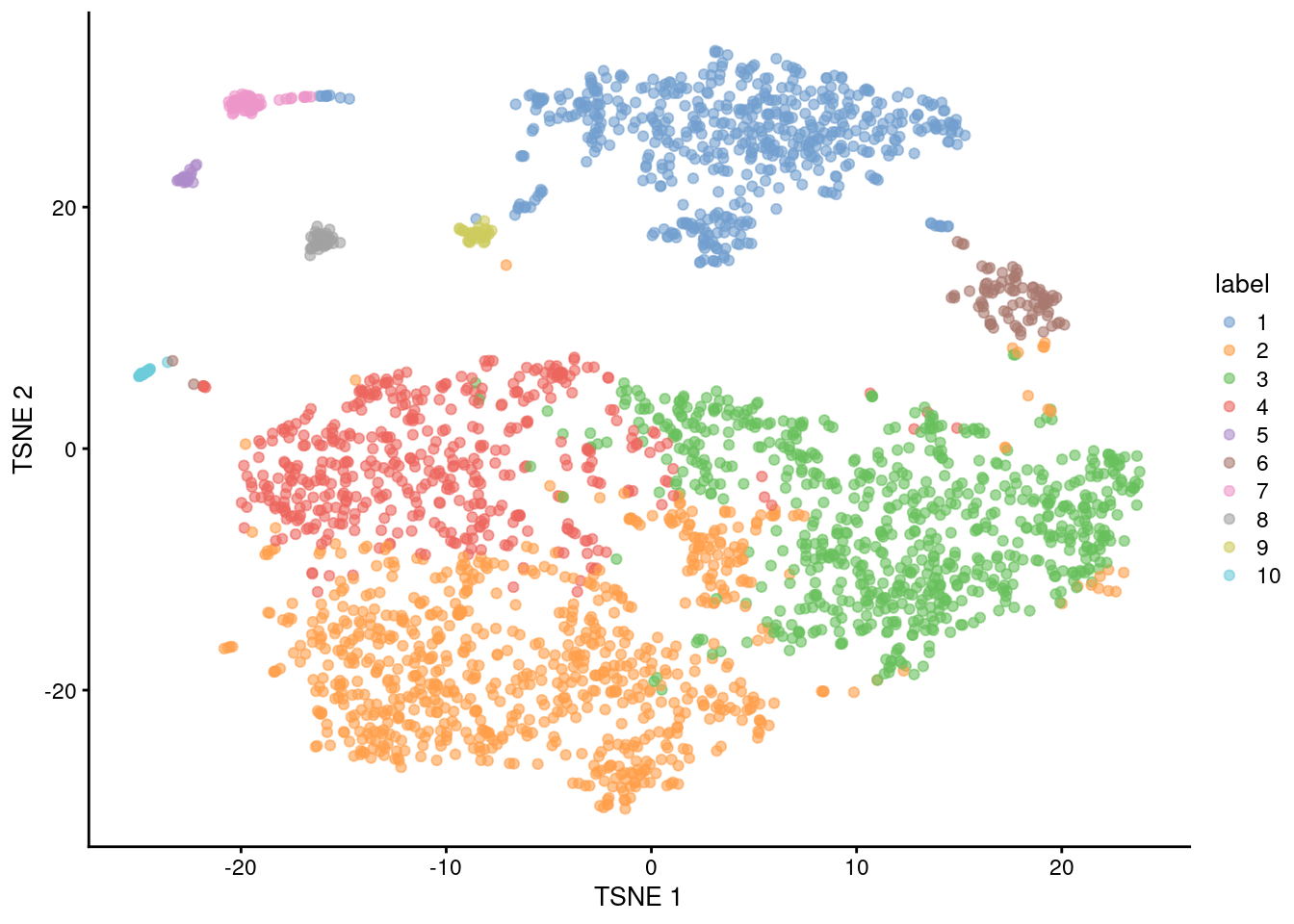
Figure 12.5: Obligatory \(t\)-SNE plot of the Bach mammary gland dataset, where each point represents a cell and is colored according to the assigned cluster.
Session Info
R version 4.1.1 (2021-08-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.3 LTS
Matrix products: default
BLAS: /home/biocbuild/bbs-3.14-bioc/R/lib/libRblas.so
LAPACK: /home/biocbuild/bbs-3.14-bioc/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_GB LC_COLLATE=C
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] BiocSingular_1.10.0 scran_1.22.0
[3] AnnotationHub_3.2.0 BiocFileCache_2.2.0
[5] dbplyr_2.1.1 scater_1.22.0
[7] ggplot2_3.3.5 scuttle_1.4.0
[9] ensembldb_2.18.0 AnnotationFilter_1.18.0
[11] GenomicFeatures_1.46.0 AnnotationDbi_1.56.0
[13] scRNAseq_2.7.2 SingleCellExperiment_1.16.0
[15] SummarizedExperiment_1.24.0 Biobase_2.54.0
[17] GenomicRanges_1.46.0 GenomeInfoDb_1.30.0
[19] IRanges_2.28.0 S4Vectors_0.32.0
[21] BiocGenerics_0.40.0 MatrixGenerics_1.6.0
[23] matrixStats_0.61.0 BiocStyle_2.22.0
[25] rebook_1.4.0
loaded via a namespace (and not attached):
[1] igraph_1.2.7 lazyeval_0.2.2
[3] BiocParallel_1.28.0 digest_0.6.28
[5] htmltools_0.5.2 viridis_0.6.2
[7] fansi_0.5.0 magrittr_2.0.1
[9] memoise_2.0.0 ScaledMatrix_1.2.0
[11] cluster_2.1.2 limma_3.50.0
[13] Biostrings_2.62.0 prettyunits_1.1.1
[15] colorspace_2.0-2 blob_1.2.2
[17] rappdirs_0.3.3 ggrepel_0.9.1
[19] xfun_0.27 dplyr_1.0.7
[21] crayon_1.4.1 RCurl_1.98-1.5
[23] jsonlite_1.7.2 graph_1.72.0
[25] glue_1.4.2 gtable_0.3.0
[27] zlibbioc_1.40.0 XVector_0.34.0
[29] DelayedArray_0.20.0 scales_1.1.1
[31] edgeR_3.36.0 DBI_1.1.1
[33] Rcpp_1.0.7 viridisLite_0.4.0
[35] xtable_1.8-4 progress_1.2.2
[37] dqrng_0.3.0 bit_4.0.4
[39] rsvd_1.0.5 metapod_1.2.0
[41] httr_1.4.2 dir.expiry_1.2.0
[43] ellipsis_0.3.2 pkgconfig_2.0.3
[45] XML_3.99-0.8 farver_2.1.0
[47] CodeDepends_0.6.5 sass_0.4.0
[49] locfit_1.5-9.4 utf8_1.2.2
[51] tidyselect_1.1.1 labeling_0.4.2
[53] rlang_0.4.12 later_1.3.0
[55] munsell_0.5.0 BiocVersion_3.14.0
[57] tools_4.1.1 cachem_1.0.6
[59] generics_0.1.1 RSQLite_2.2.8
[61] ExperimentHub_2.2.0 evaluate_0.14
[63] stringr_1.4.0 fastmap_1.1.0
[65] yaml_2.2.1 knitr_1.36
[67] bit64_4.0.5 purrr_0.3.4
[69] KEGGREST_1.34.0 sparseMatrixStats_1.6.0
[71] mime_0.12 xml2_1.3.2
[73] biomaRt_2.50.0 compiler_4.1.1
[75] beeswarm_0.4.0 filelock_1.0.2
[77] curl_4.3.2 png_0.1-7
[79] interactiveDisplayBase_1.32.0 statmod_1.4.36
[81] tibble_3.1.5 bslib_0.3.1
[83] stringi_1.7.5 highr_0.9
[85] bluster_1.4.0 lattice_0.20-45
[87] ProtGenerics_1.26.0 Matrix_1.3-4
[89] vctrs_0.3.8 pillar_1.6.4
[91] lifecycle_1.0.1 BiocManager_1.30.16
[93] jquerylib_0.1.4 BiocNeighbors_1.12.0
[95] cowplot_1.1.1 bitops_1.0-7
[97] irlba_2.3.3 httpuv_1.6.3
[99] rtracklayer_1.54.0 R6_2.5.1
[101] BiocIO_1.4.0 bookdown_0.24
[103] promises_1.2.0.1 gridExtra_2.3
[105] vipor_0.4.5 codetools_0.2-18
[107] assertthat_0.2.1 rjson_0.2.20
[109] withr_2.4.2 GenomicAlignments_1.30.0
[111] Rsamtools_2.10.0 GenomeInfoDbData_1.2.7
[113] parallel_4.1.1 hms_1.1.1
[115] grid_4.1.1 beachmat_2.10.0
[117] rmarkdown_2.11 DelayedMatrixStats_1.16.0
[119] Rtsne_0.15 shiny_1.7.1
[121] ggbeeswarm_0.6.0 restfulr_0.0.13 References
Bach, K., S. Pensa, M. Grzelak, J. Hadfield, D. J. Adams, J. C. Marioni, and W. T. Khaled. 2017. “Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing.” Nat Commun 8 (1): 2128.